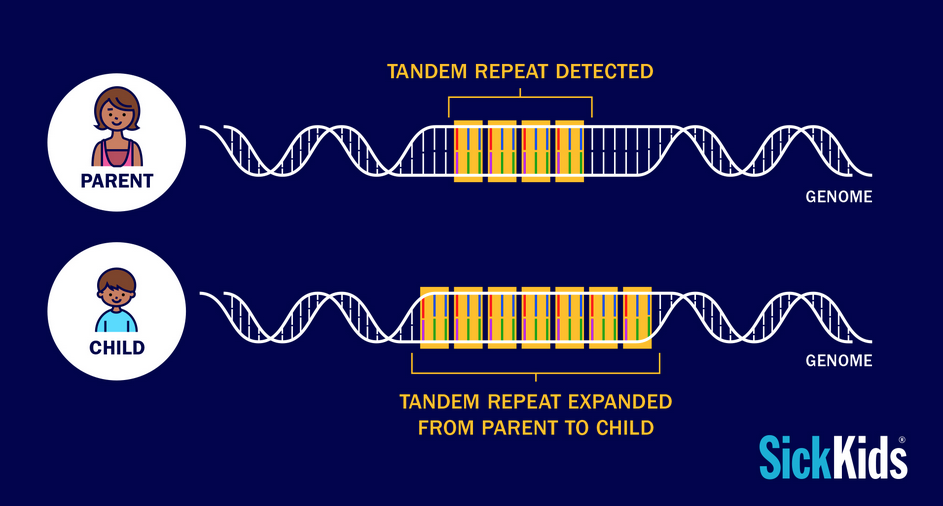
Tandem Repeat Expansions
Tandem repeats (TRs) are a class of DNA sequence where two or more bases are repeated immediately adjacent to one another. Expansions of these repetitive regions are suspected to contribute to the genetic etiology of complex disorders.
Short-read whole-genome sequencing (WGS) has trouble characterizing these regions due to ambiguous mapping. We are developing and implementing new technologies and computational algorithms to perform genome-wide screens of repetitive DNA. Our aims are to genotype TRs on large genomic datasets as well as determine the role, and characterize the mechanism, of TR variation.
Whole Genome Sequencing Analysis of Neurological Disorders
Many neurological disorders have a complex genetic origin, meaning the same, single gene cannot explain the phenotype for most cases. We carry out WGS analysis to investigate different forms of rare genetic (including single nucleotide, copy number, and TR) variation in neurological disorders, such as autism spectrum disorder, movement disorders, epilepsy, and schizophrenia.
Participate in our research!
We are actively recruiting participants with unexplained pediatric-onset movement disorders for a whole genome sequencing study. Check out our study poster or expand the section below to learn more.
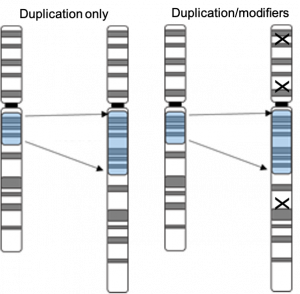
Genetic Modifiers
Pathogenic genetic variants often exhibit incomplete penetrance and variable expressivity. Additional genetic factors (e.g., single nucleotide variants, copy number variants, structural variants, and TR expansions), which may act as modifiers, are sometimes present in affected individuals. We are studying this phenomenon in a number of disorders, including the 7q11.23 duplication syndrome, the 22q11.2 deletion syndrome, and in TR expansion related diseases such as motor neuron disease.