Single cell RNA sequencing (scRNA-seq), combined with multi-omics approaches, enables simultaneous profiling of gene expression, protein, and epigenetic features at single cell resolution. This integrated analysis provides powerful insights into cellular heterogeneity, function, and lineage dynamics in both fundamental and translational research.
Single cell multi-omics technologies are broadly applicable across immunology, oncology, neurobiology, and genetic disease research areas.
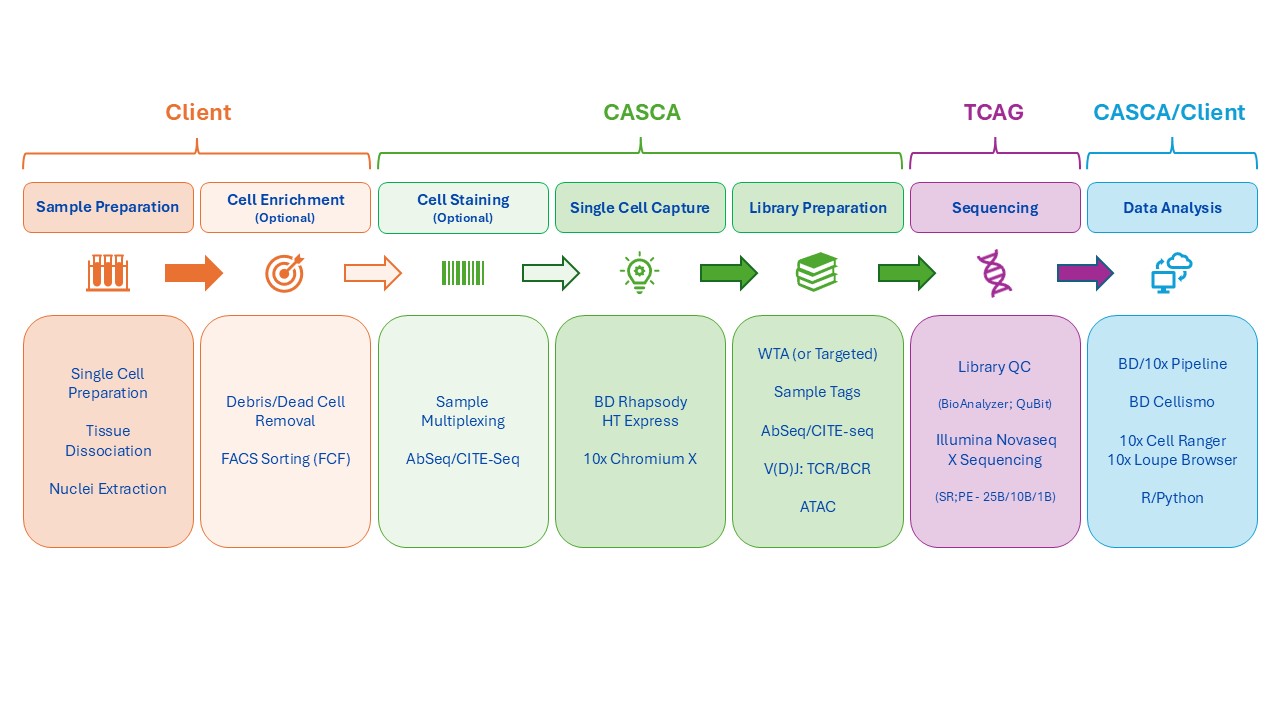
CASCA offers the BD Rhapsody HT Xpress and 10x Genomics Chromium X single cell multi-omics platforms. We offer consultation for experimental design, platform and assay selection and sample preparation optimization. We perform single cell antibody labeling (optional), cell capture and library preparation. Sequencing is performed in partnership with The Centre for Applied Genomics (TCAG) at SickKids. We also provide basic downstream data analysis to assist with initial interpretation and quality assessment.
To learn more or to obtain a quote for your project: submit a “Single Cell Multi-omics Experimental Design Consultation” service request (listed in the Single Cell Multi-omics category) in iLab.
When choosing your platform, consider your specific experimental goals, sample type (s), and desired data output. For additional details on platform specifications, assay capabilities, and updated protocols, visit the vendor websites:
- BD Biosciences: https://scomix.bd.com/hc/en-us
- 10x Genomics: https://www.10xgenomics.com/platforms/chromium
——————————————-
See table below for overview and comparison of the BD Rhapsody HT Xpress and 10x Genomics Chromium X
| Platform Overview | BD Rhapsody HT Xpress | 10x Chromium X |
| Single Cell Capture | In microwells with barcoded magnetic beads (gravity); no clogging | In oil-emulsion droplets with barcoded gel beads (microfluidics) |
| Capture Options | 8 lanes per cartridge; charged by lanes used | 8 wells per chip; charged for whole chip |
| Cell Type Compatibility | Effective for most cell types; gentle capture preserves fragile cells | Effective for most cell types; less optimal for fragile cells |
| Pre-library Capture Validation/QC | Integrated imaging quantifies wells with single viable cells + bead | None |
| – | – | – |
| Workflow Options | BD Rhapsody HT Xpress | 10x Chromium X |
| Sample Multiplexing | Up to 24 samples per lane | On-chip multiplexing (3’and 5′); in-line multiplexing (Flex) |
| Multiplexing Method | Oligo-tagged antibodies bind ubiquitous surface or nuclear markers | Lipid-modified oligos integrate into cell membranes |
| Maximum # cells to load | 100,000 per lane, 800,000 per cartridge | 20,000 per sample, 160,000 per chip |
| – | – | – |
| Assay Types | BD Rhapsody HT Xpress | 10x Chromium X |
| mRNA WTA | Whole Transcriptome Analysis (WTA): add link | Universal 3′ Gene Expression |
| Targeted Gene Expression | ~400 human or mouse genes: pre-configured or customizable | NA |
| Protein Expression | AbSeq oligo-antibodies – surface & intracellular (all assays) | CITE-seq oligo-antibodies – surface (all assays) & intracellular (Flex assay only) |
| Immune Profiling | WTA+ full-length TCRA, TCRB, TCRG, TCRD, IGHM, IGK, IGL | 5′ gene expression + full-length TCRA, TCRB, IGHM, IGK, IGL |
| ATACseq (nuclei) | WTA + chromatin accessability, optional sample multiplexing (up to 6 samples) | Chromatin accessability only or 3′ gene expression + chromatin accessability |
| Fixed Cell Assay | NA | Flex: Probe-based gene expression; optional cell surface and/or intracellular protein |
| – | – | – |
| Data Analysis | BD Rhapsody HT Xpress | 10x Chromium X |
| Pre-Processing | BD pipeline (SevenBridges) & | 10x pipelines (e.g., Cell Ranger, Loupe Browser) |
| Analysis | BD Cellismo data visualizaton tool, R/Python | R/Python |