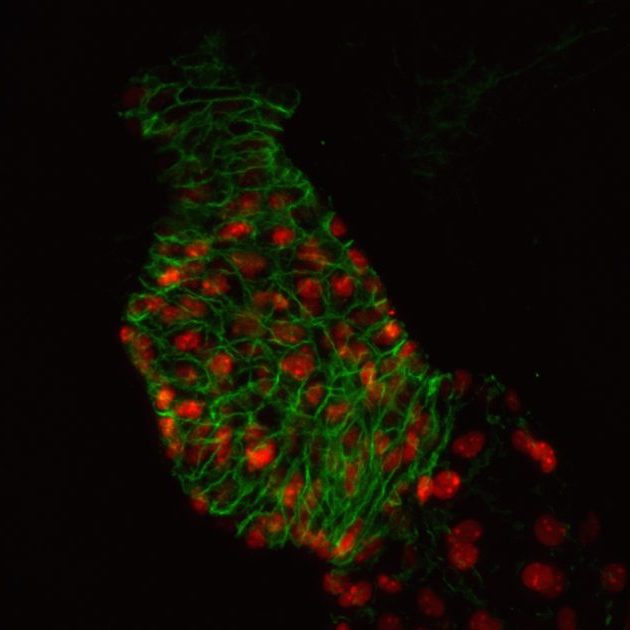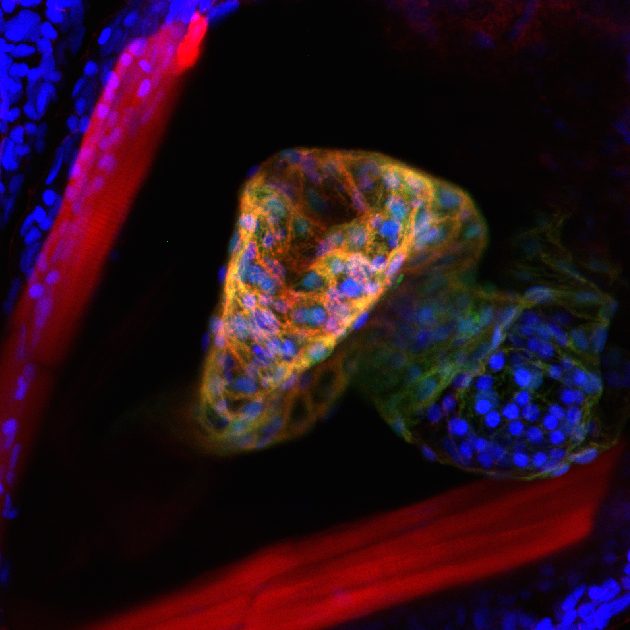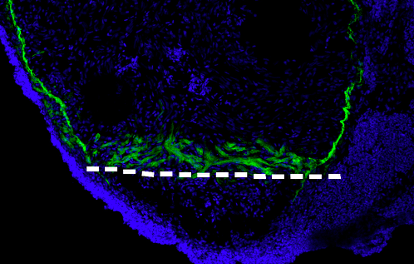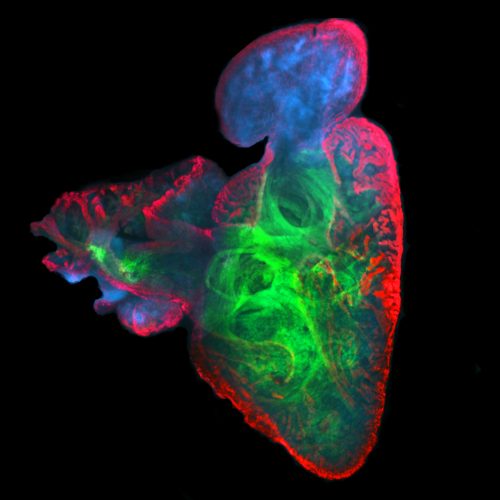
We use a number of approaches to carry out our research, including creation of numerous mutant and transgenic lines to examine gene function and image cells as they build the heart in real time. In collaboration with Dr. Michael Wilson at SickKids, we use sophisticated genomics approaches to discover genetic elements key to cardiac development and disease. By defining what is conserved between zebrafish and human, a gap that spans over 400 million years of evolution, we are starting to define the critical drivers of heart development and disease. Combined with the explosion of human genome sequences that are available, we aim to pinpoint the key genetic events that lead to cardiovascular disease, and that can be exploited for therapies. Further, the embryonic heart development program is re-deployed during adult zebrafish cardiac regeneration. We are therefore interested in studying the ability of key developmental gene regulatory networks to drive a regenerative response.
In the long term, this knowledge will be used to devise therapies based on activating regenerative pathways to heal the damaged heart, and using zebrafish embryos to screen for potential drugs to alleviate the burden of congenital heart defects and other cardiovascular diseases, including cerebral cavernous malformations (CCMs). Discovering how zebrafish cardiac progenitors are made and develop will therefore drive future efforts to repair what is at present irreparable heart disease.
Trainees working on this project:
Nathan Stutt, Maria Fahim, Cherry Liu
We have a major interest in the earliest events of heart development that regulate how cardiac progenitors, the building blocks of the heart, are first specified and diversified. Our early work with the grinch mutant has shown that function of the Apelin receptor GPCR (Aplnr) is essential for cardiac development, with aplnr/a/b mutants being heartless. This appears to be due to defects in the cardiac lineage at the earliest events of development, as the gastrulation is being initiated. To address this, we have collaborated with the lab of Dr. Michael Wilson at SickKids to undertake single cell RNAseq and ATACseq analysis of the initial stages of cardiac development in both wild type and heartless (aplnra/b and gata5/6 loss-of-function) models. We have found that regulation of gata5 expression is a key event in diversification of cardiopharyngeal lineages, a mechanism that we are actively studying. This work will lead to the key gene regulatory networks that govern the earliest events of cardiac lineage specification.
Subsequent to cardiac lineage commitment, cardiac progenitor cells must be diversified to form the various components of the heart. Our lab was amongst the first to demonstrate that zebrafish heart development occurs via two waves of addition of cells from the so-called first and second heart fields (FHF and SHF), as is true in mammals. Subsequently, these cells differentiate to form the structures of the heart. We are utilizing a comparative genomics approach to identify genes and gene regulatory elements (enhancers) conserved in cardiac expression/activity from zebrafish to human. These conserved elements are being examined for activity in transgenic and mutant models we are generating. We are in particular prioritizing enhancers with ultrarare variants in patients with congenital heart disease. Long-term goals include creating zebrafish models of congenital heart disease for functional and small molecule therapeutic screens, annotating non-coding variants associated with human disease, and identifying genes and enhancers that may play roles in cardiac regeneration.
Trainees working on this project:
None currently
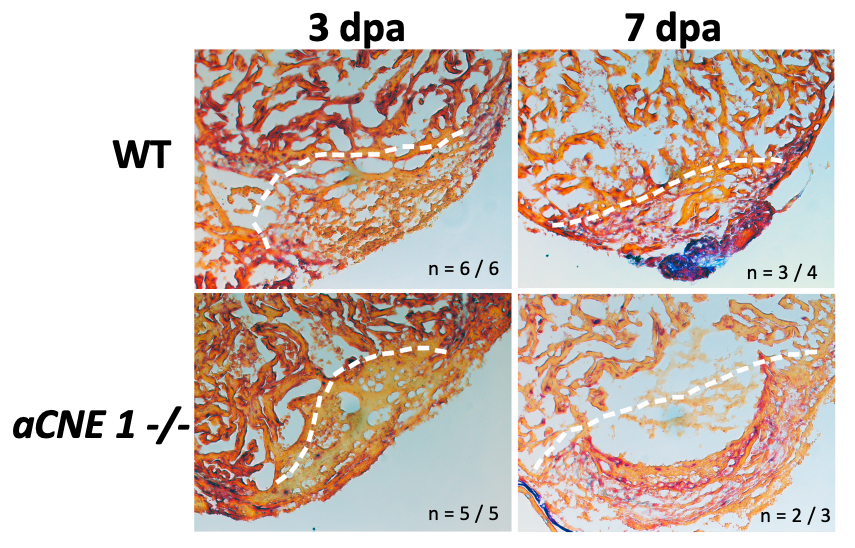
Unlike the case in mammals, the adult zebrafish heart has a remarkable regenerative capacity. We have generated a number of transgenic and mutant lines based on enhancers we have found to be active during cardiac development and conserved in function between zebrafish and humans. Intriguingly, we have identified several that are activated upon cardiac injury, and are currently examining their function in this process. Combined with our work in cardiac development, we aim to identify key molecular events that can be used to drive cardiac repair in the human heart.
- Early Cardiac Lineages
Trainees working on this project:
Nathan Stutt, Maria Fahim, Cherry Liu
We have a major interest in the earliest events of heart development that regulate how cardiac progenitors, the building blocks of the heart, are first specified and diversified. Our early work with the grinch mutant has shown that function of the Apelin receptor GPCR (Aplnr) is essential for cardiac development, with aplnr/a/b mutants being heartless. This appears to be due to defects in the cardiac lineage at the earliest events of development, as the gastrulation is being initiated. To address this, we have collaborated with the lab of Dr. Michael Wilson at SickKids to undertake single cell RNAseq and ATACseq analysis of the initial stages of cardiac development in both wild type and heartless (aplnra/b and gata5/6 loss-of-function) models. We have found that regulation of gata5 expression is a key event in diversification of cardiopharyngeal lineages, a mechanism that we are actively studying. This work will lead to the key gene regulatory networks that govern the earliest events of cardiac lineage specification.
- Conserved Enhancers in Heart Development and Disease
Subsequent to cardiac lineage commitment, cardiac progenitor cells must be diversified to form the various components of the heart. Our lab was amongst the first to demonstrate that zebrafish heart development occurs via two waves of addition of cells from the so-called first and second heart fields (FHF and SHF), as is true in mammals. Subsequently, these cells differentiate to form the structures of the heart. We are utilizing a comparative genomics approach to identify genes and gene regulatory elements (enhancers) conserved in cardiac expression/activity from zebrafish to human. These conserved elements are being examined for activity in transgenic and mutant models we are generating. We are in particular prioritizing enhancers with ultrarare variants in patients with congenital heart disease. Long-term goals include creating zebrafish models of congenital heart disease for functional and small molecule therapeutic screens, annotating non-coding variants associated with human disease, and identifying genes and enhancers that may play roles in cardiac regeneration.
- Cardiac Regeneration
Trainees working on this project:
None currently
Unlike the case in mammals, the adult zebrafish heart has a remarkable regenerative capacity. We have generated a number of transgenic and mutant lines based on enhancers we have found to be active during cardiac development and conserved in function between zebrafish and humans. Intriguingly, we have identified several that are activated upon cardiac injury, and are currently examining their function in this process. Combined with our work in cardiac development, we aim to identify key molecular events that can be used to drive cardiac repair in the human heart.