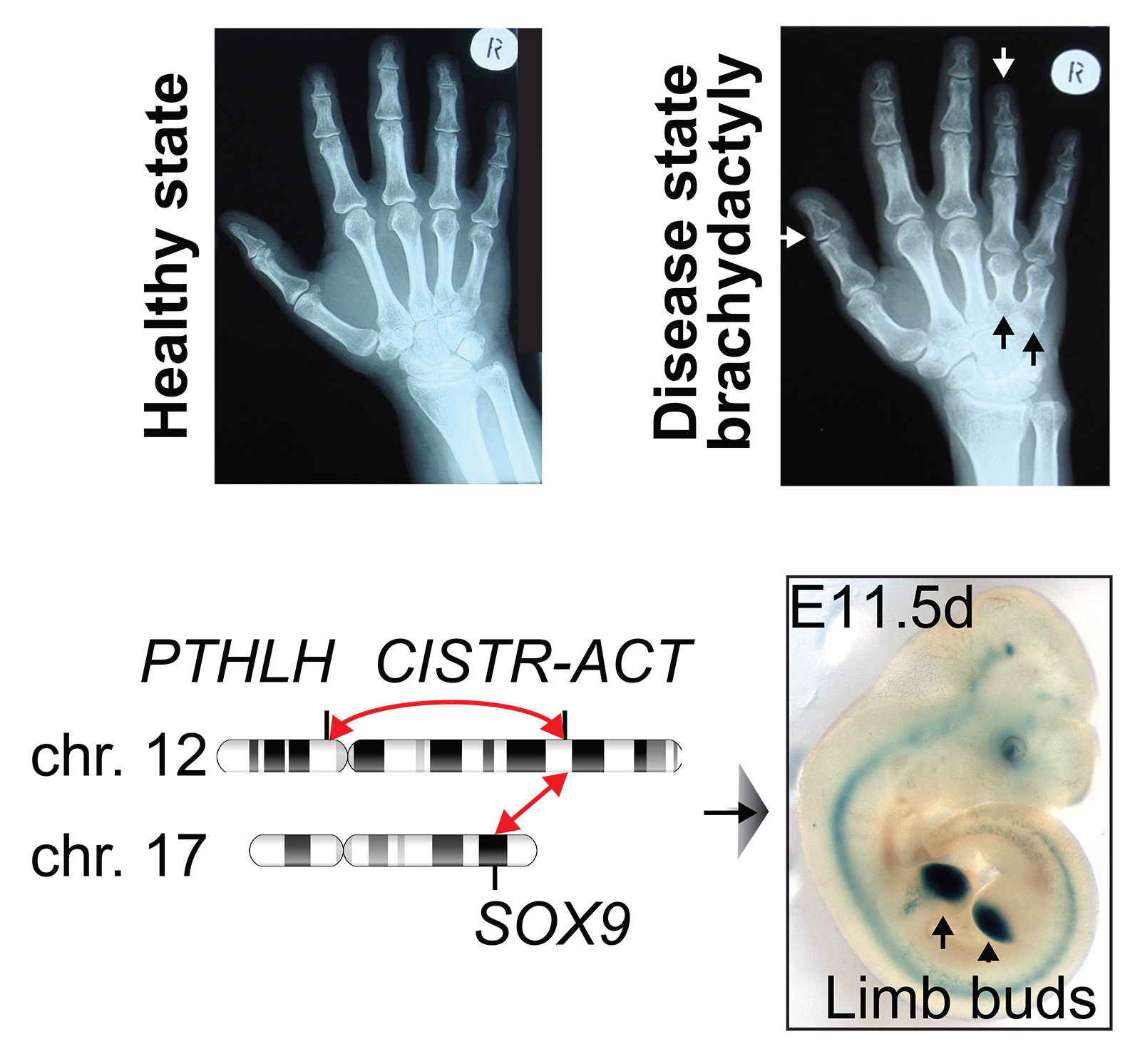
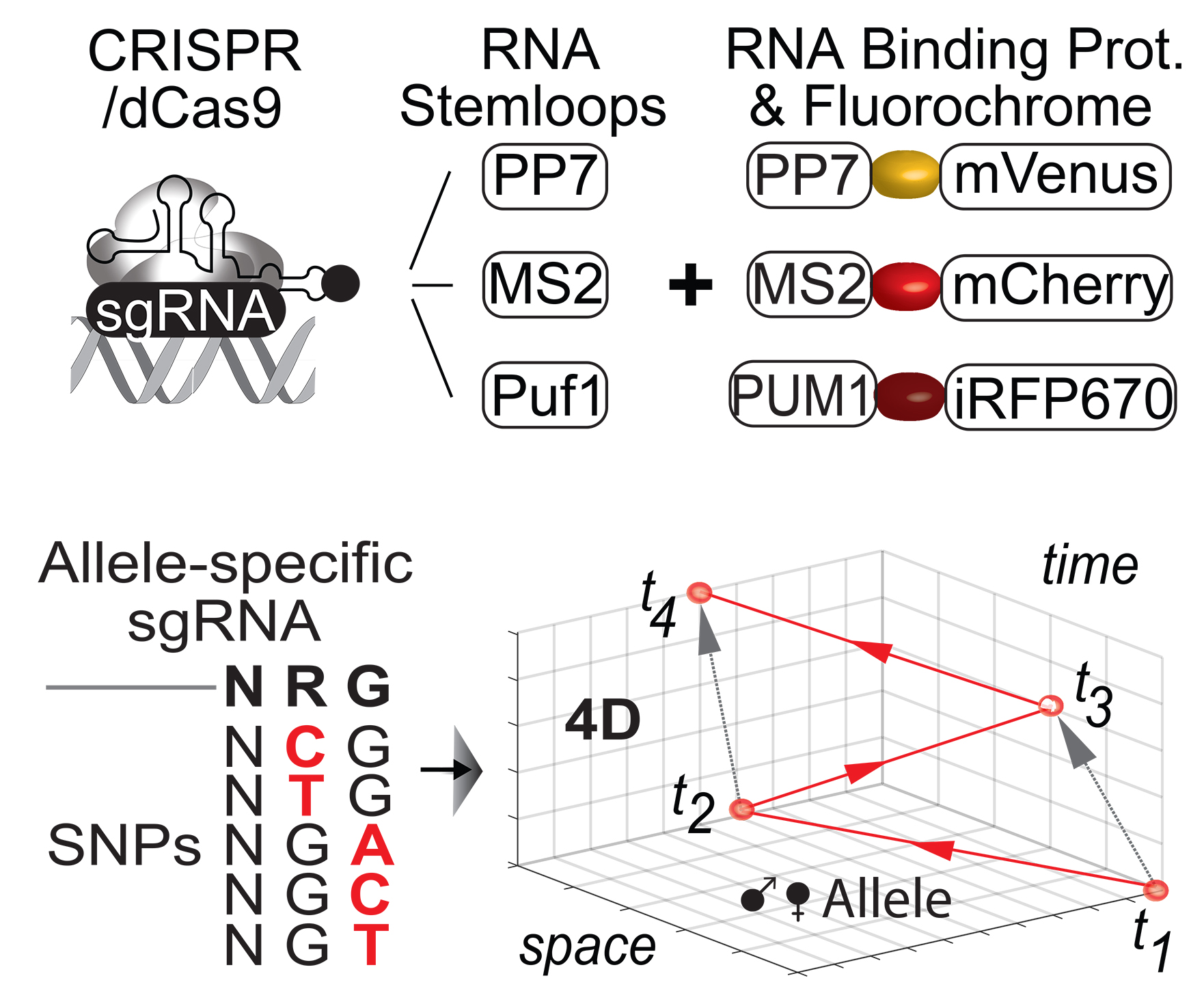
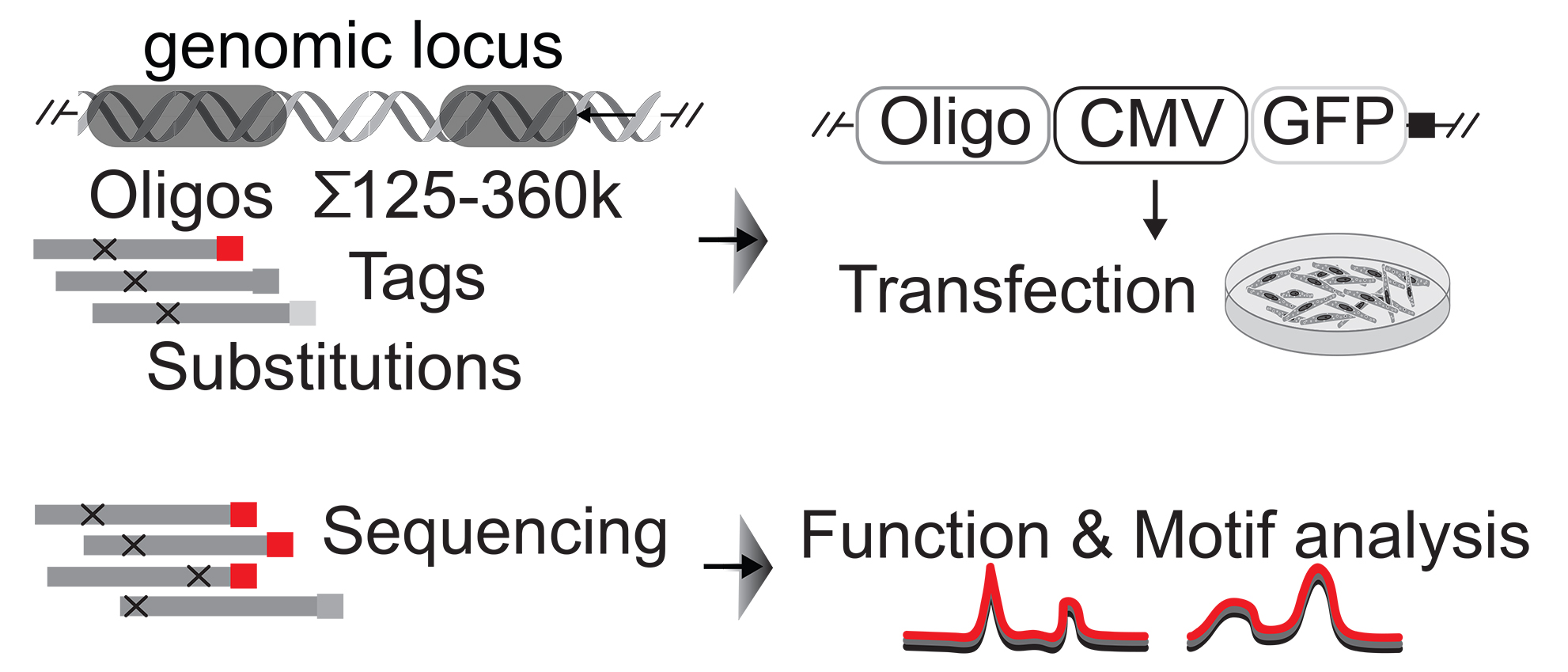
Genome organization is important for the regulation of genes and cell fate determinations. For gene expression to occur, regulatory elements come into physical proximity of genes. Rearranging these gene regulatory networks and genome organization alters developmental programs and causes rare diseases and cancer.
Various disease-associated protein-coding genes have been identified. However, coding genes represent only ~2% of the transcribed human genome, while > 95% consists of transcribed non-coding DNA. The majority of disease-relevant genetic variations and mechanisms reside in the non-coding genome, including long non-coding RNAs (lncRNAs) which do not encode for proteins. Intensive research demonstrates that lncRNAs and noncoding DNA are integral to the function of cells, particularly in the control of gene activity and in the maintenance of genome organization, a discovery revolutionizing our understanding of biology and of pathogenesis.
To better understand biology and etiology of disease, we need to determine how the non-coding genome functions and how it impacts normal and disease states. The Maass Lab studies molecular mechanisms associated with the non-coding genome.