NET Formation Pathways
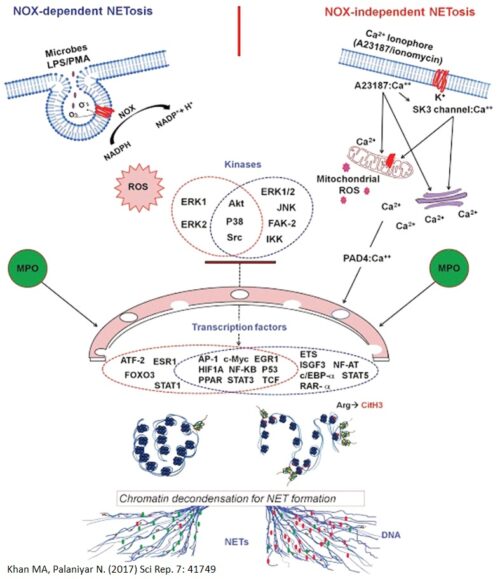
NETosis or NET formation can occur via 2 different pathways depending on the agonist used, and are differentiated by whether certain enzymes, specifically NADPH Oxidase 2 (NOX2) are used.
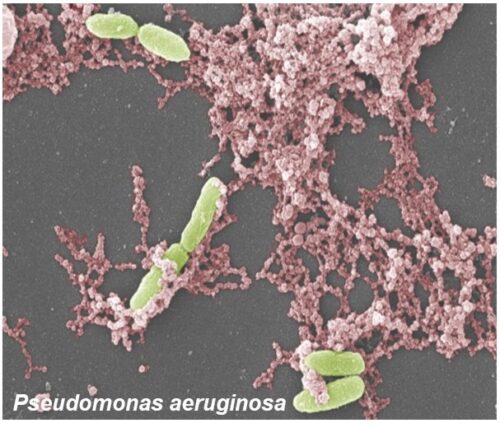
Lipopolysaccharide (LPS), a component of gram-negative bacteria, and Phorbol 12-myristate 13-acetate (PMA), a synthetic compound, are stimulators of the NOX-dependent Pathway. These agonists will stimulate NOX2 to produce large amounts of reactive oxidative species (ROS) which contain free radicals (The stuff that antioxidants neutralize).
Whereas the influx of calcium ions into the neutrophils, whether caused by the administration of drugs (Ex. calcium ionophores) or activation of transporter proteins (Ex. SK3 channels) promote the NOX-independent pathway to produce NETs faster than the other pathway. The influx of calcium ions into the neutrophil will encourage the mitochondria to produce ROS. These calcium ions also bind with PAD4, initiating histone citrullination, a process that facilitates chromatin decondensation which unravels the DNA within the cell nucleus.
Both of these ROS will activate different sets of kinases, and disintegrate the membranes of the nucleus and granules. The disintegration of the membranes will allow proteins, granules, and enzymes such as Myeloperoxidase and Neutrophil Elastase to interact with and unravel the DNA (to cleave the histones, or facilitate chromatin decondensation). These decondensed DNA at this point are decorated with granular contents and are released out of the neutrophils as NETs.
An important discovery of our lab was the role of transcriptional firing in NETosis. We found that NETosis-specific kinase cascades activate transcription factors, starting transcription at multiple loci at all chromosomes to facilitate chromatin decondensation (An important step in creating RNA and later on, proteins). When we inhibit transcription, both the NOX-dependent and NOX-independent pathways were suppressed. However, inhibition of translation (Process that makes proteins) did not affect NETosis.
ROS Induces DNA Repair in NETosis
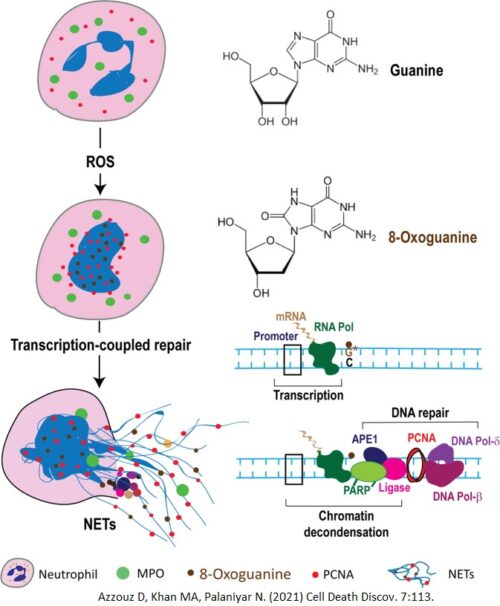
A new and exciting discovery! We discovered that ROS induces NETosis by initiating DNA repair. When ROS interacts with the chromatin in the nucleus, they perform extensive DNA damage, oxidizing the guanine residues in the DNA, creating 8-Oxoguanine (Markers of DNA lesions). When transcription occurs, such as when NETosis causes genome-wide transcription, the transcription machinery will stall at the sites of DNA lesions and signal other proteins to initiate DNA repair.
The DNA repair machinery assemble at the sites of DNA damage, and in the process of repairing, decondenses the chromatin. The DNA-unwinding capabilities of these DNA repair proteins are key drivers of NETosis.
Frequently Used Techniques
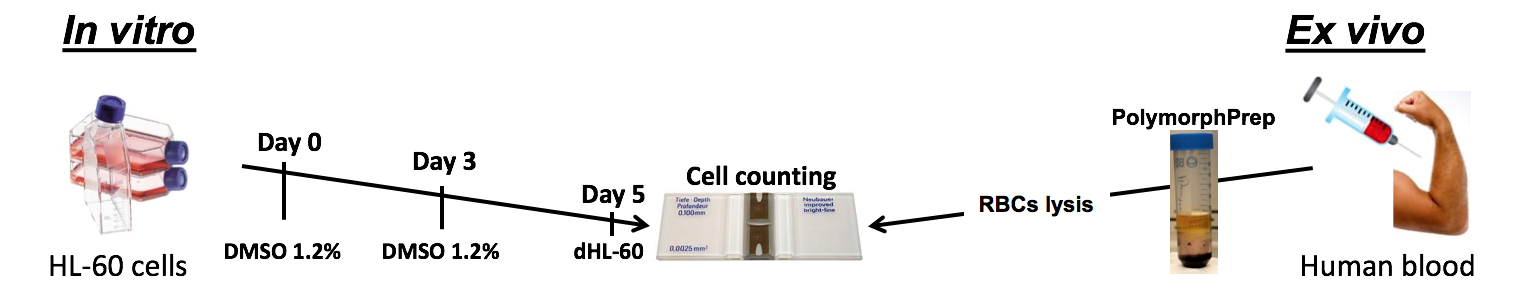
Below is a diagram outlining techniques we use regularly in our experiments regarding NETs. This work can be done with either HL-60 cells (in vivo) that can be differentiated into neutrophil-like cells using DMSO, or primary neutrophils obtained from volunteers (ex vivo).
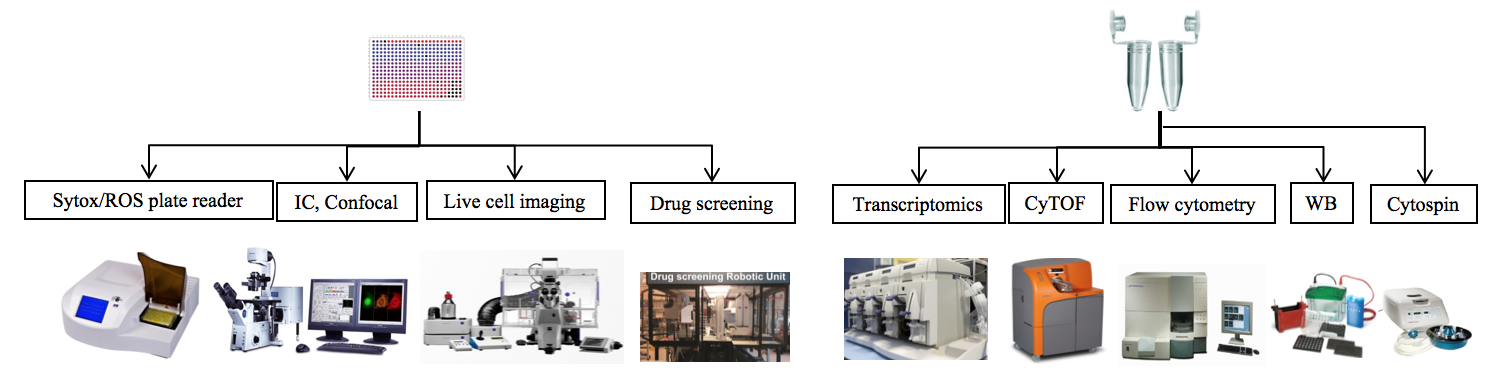
Once the cells are properly isolated, they will be stimulated by a variety of agonists. These include: PMA, A23187, ionomycin, LPS, P. a, S. a, PAF, Pyocyanin, IC, and HxA3. After they are stimulated, the cells will follow two different paths. They can be pipetted into 96-well plates for to be imaged, to have their ROS levels measured, or have cell viability measured among other things. They could also be left in the microtube so that other aspects of the cells can be measured such as protein production, and gene transcription. Possible methods are shown in the image below.
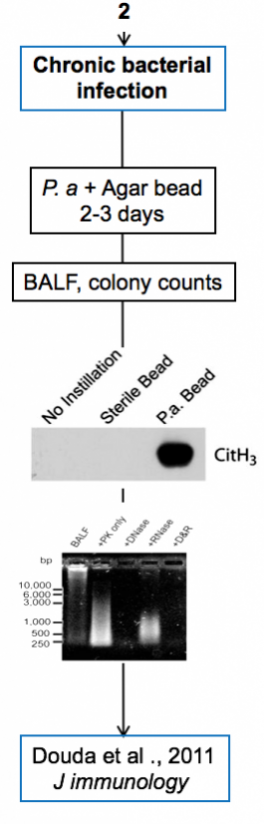
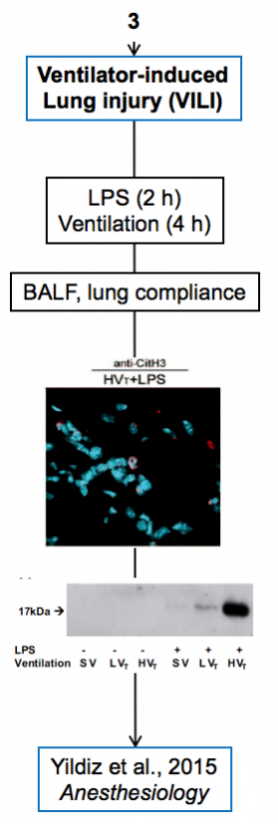
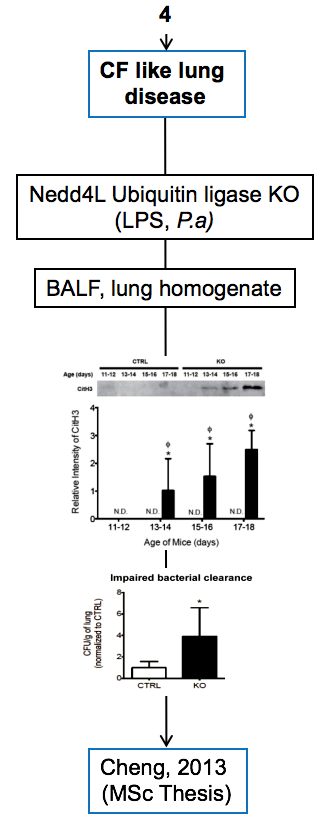
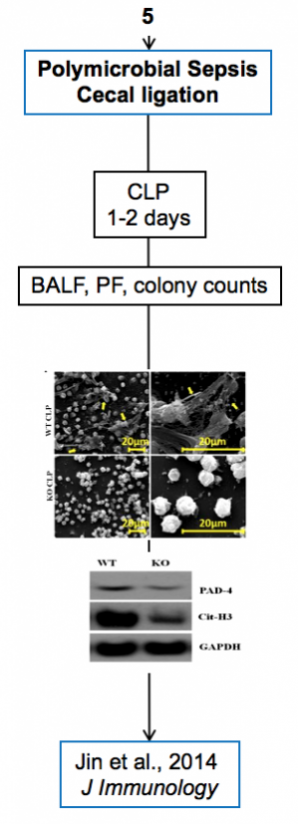
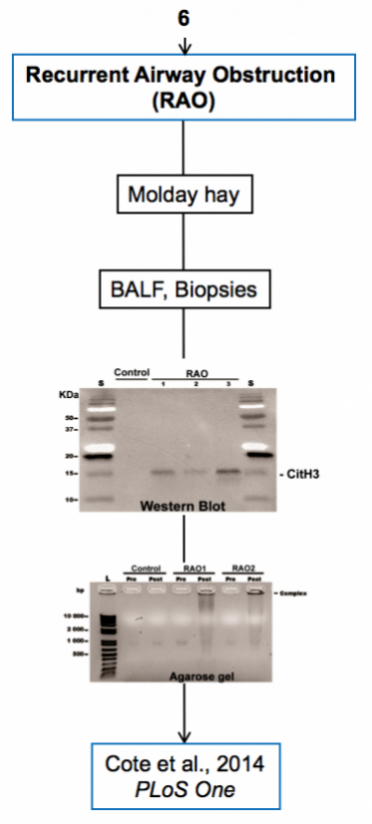
Experimental models
Below are diagrams that depict the various experimental models used in our experiments. The majority of our models are in mice, though we do use a single horse model. The diagrams show the type of model, briefly how they are created, data depicting their use, and publications in which they are utilized.