iPS Cells to Study Human Disease
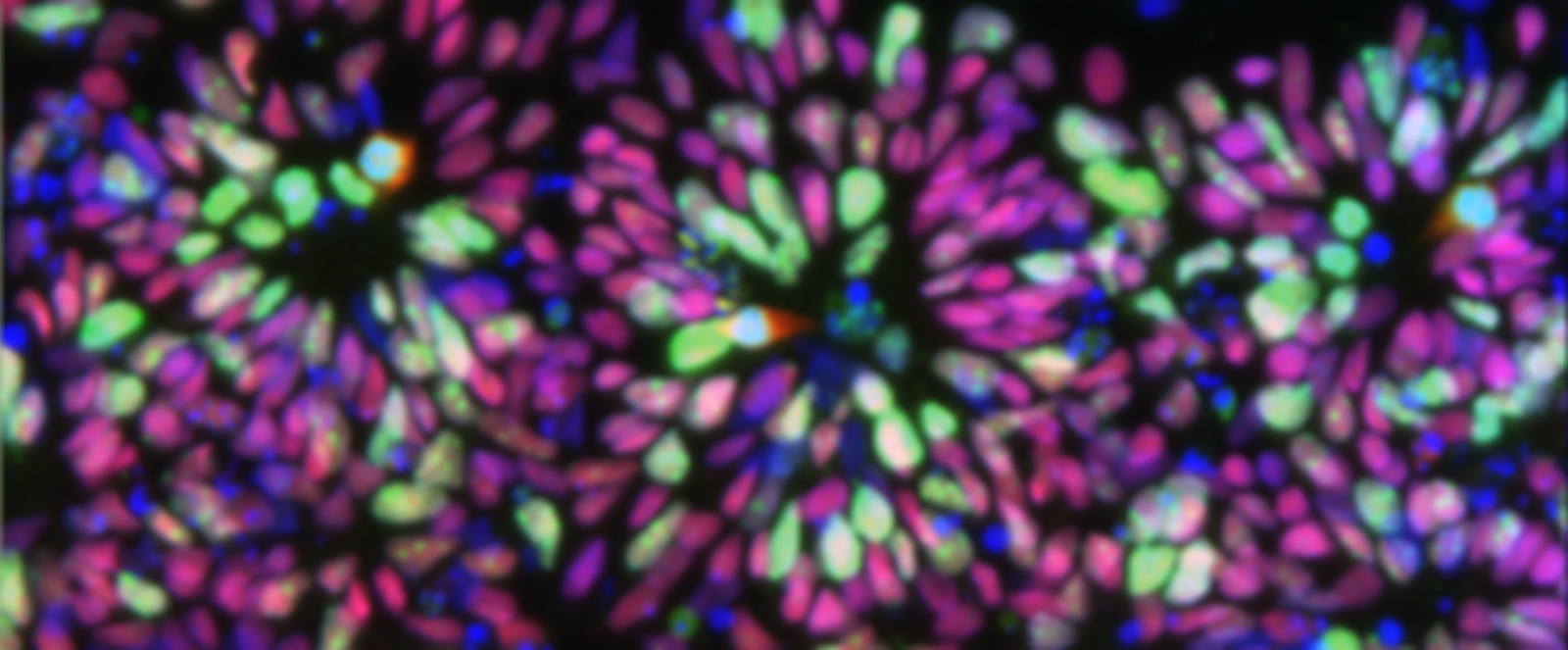
Induced Pluripotent Stem (iPS) cells can make any cell type of the body and are generated by inducing skin cells to change into stem cells in culture. This cell induction is called “reprogramming” and is performed by delivering a combination of four genes that normally control the stem cell state. We made mouse and human iPS cells using these four genes, and developed a novel EOS lentivirus vector that signals when the skin cells have become iPS cells. This technology is in use to study mechanisms of human disease by inducing patient skin cells to become iPS cells, and then differentiating the iPS cells into the affected cell type for in vitro tests.
Rett syndrome and autism spectrum disorder
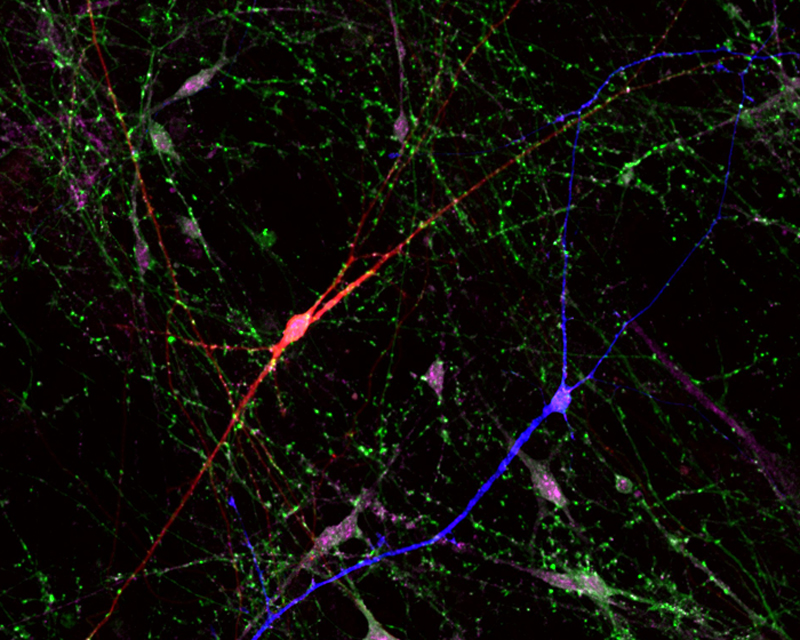
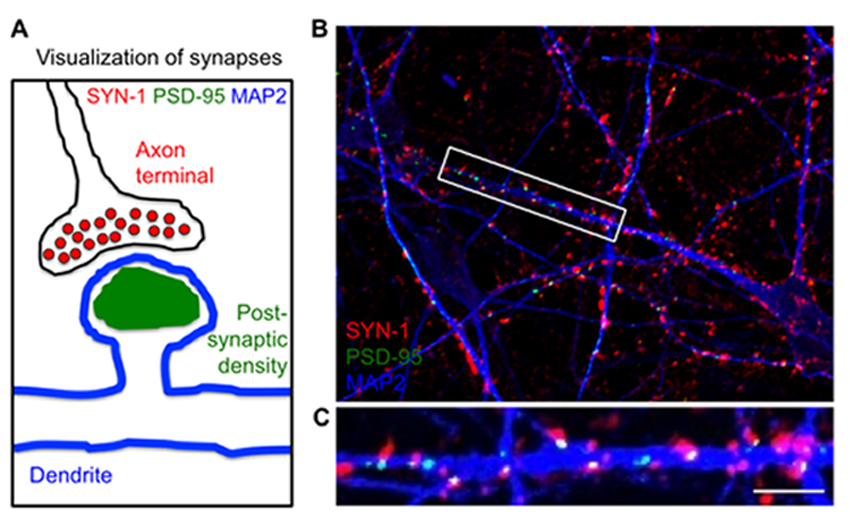
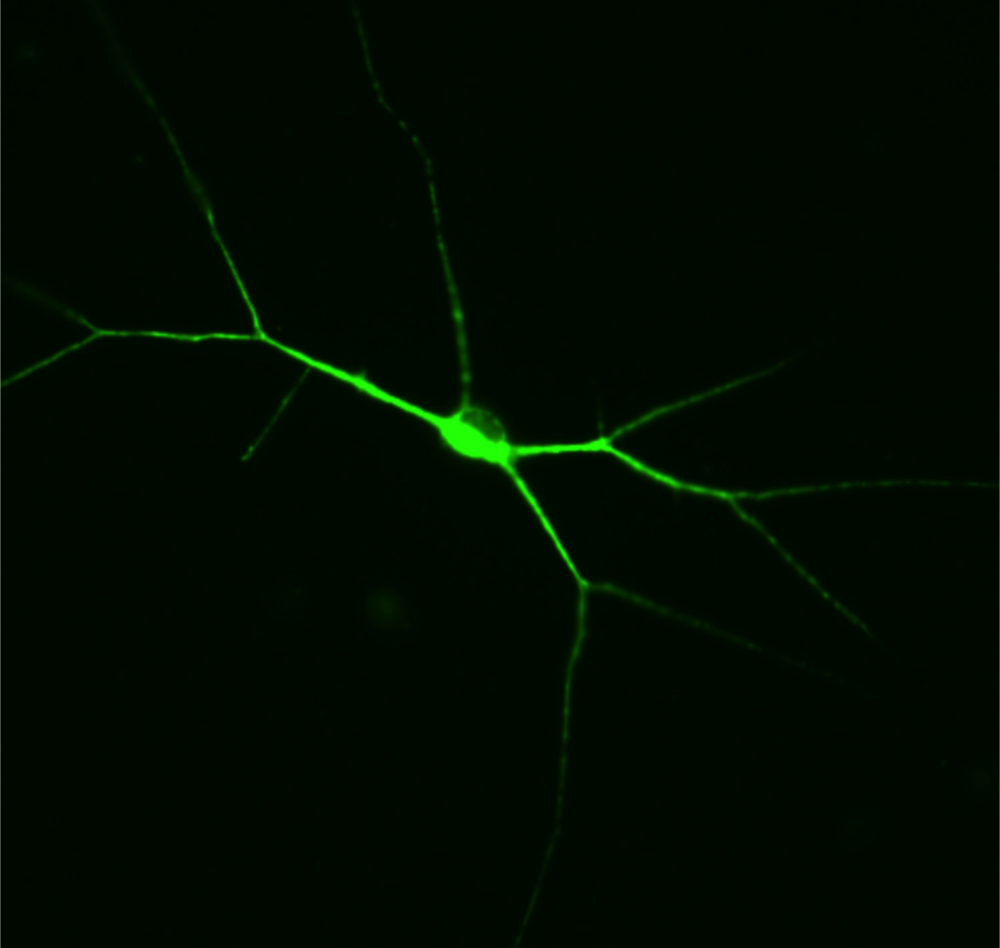
Our primary interest for modeling human disease using iPS cell technology is aimed toward Rett Syndrome (RTT). It is caused by defects in the X-linked MECP2 gene that specifically affects girls with devastatingly early onset leading to profound developmental challenges and autistic behavior. RTT is an outstanding candidate for disease studies using iPS cells because it is not ethically possible to obtain neurons from the brains of these individuals. To study the defects in neurons with RTT disease, we generated mouse and human iPS cells using skin cells from RTT knockout mice, RTT patients and healthy controls. We differentiated these iPS cells into neurons to identify abnormal functions of RTT neurons in culture with the Salter lab and rescued the RTT neuron abnormalities by delivery of a normal MECP2 gene. Overall, these studies determine the functional defects in RTT nerve cells derived from patients to reveal mechanisms of disease. Together with the Scherer lab we generated iPS cells from subjects with autism spectrum disorders (ASD) and knocked out ASD associated genes using CRISPR-Cas9 genome editing. We are currently differentiating these lines into neurons for phenotyping using multi-electrode arrays, and examining the neural circuitry changes with the Martinez lab at Western University. Importantly, these iPS cells can now be used to screen for new patient-specific drugs to treat RTT and ASD.
The RTT and ASD studies are funded by CIHR, Colonel Harland Sanders Rett Syndrome Research Fund, Simons Foundation Autism Research Initiative, and McLaughlin Centre. The ASD research was initiated with funds from the NIH and the Stem Cell Network.
Williams Beuren syndrome and Cardiomyopathy
Williams Beuren syndrome (WBS) is caused by a deletion that removes 26 genes from Chromosome 7q11.23. The major effects are cardiovascular malformations due to heterozygous deletion of the ELASTIN gene resulting in stenosis that must be surgically corrected. Deletion of other genes within the critical region leads to neurodevelopmental consequences including intellectual disability. Together with the Mital lab, we reprogrammed skin cells from WBS patients and differentiated them into vascular smooth muscle cells. In comparison to healthy controls, the WBS cells over-proliferated and were less effective at differentiating into mature functional cells. Preliminary studies suggested that the approved cancer drug rapamycin may be repurposed to partially correct the vascular defects, and we are currently using the iPS cell derived smooth muscle cells for drug testing and screening. We are expanding these studies to include cells with ELASTIN mutations or with duplications of the WBS critical region. These WBS studies were funded by CIHR.
The Mital lab is also discovering genetic variants in patients with Cardiomyopathies (CMP). We generate iPSC from the patients or introduce patient-specific variants into healthy control iPSC. To model CMP, we differentiate the stem cells into beating ventricular cardiomyocytes in vitro and the Mital lab phenotypes them using xCELLigence contractility and other assays. Variant specific phenotypes will then be screened for rescue by pre-approved targeted drugs. These CMP studies are funded by the Ted Rogers Centre for Heart Research.
Healthy controls for disease modeling and gene editing
The Personal genome Project Canada (PGP-C) has shown that all healthy individuals harbour heterozygous pathogenic genetic variants, suggesting that all healthy control cell lines have a genetic burden of predisposing mutations. We have generated iPSC from 3 individuals in the PGP-C and shown that they efficiently differentiate in vitro into cells from all three germ layers. Functional phenotyping has been completed on many of these cell types including cortical neurons, cardiomyocytes and hepatocytes. We will make these lines available for the stem cell community as a resource, and develop guidelines to identify the most appropriate healthy control lines to use for tissue specific disease modeling or gene-editing studies. This work is funded by the McLaughlin Centre, Ted Rogers Centre for Heart Research and the POND network.