Our research focus spans from gene discoveries to the development of personalized therapies for rare inherited diseases. There are more than 7000 rare diseases worldwide, where children make up 50% of the affected population. Among all the rare diseases, only 5% have treatments. We are part of the effort in developing therapies mediated through gene editing and gene modulation strategies for these hereditary rare diseases.
Projects
LAMA2 Congenital Muscular Dystrophy (LAMA2-CMD)
LAMA2-CMD is an autosomal recessive neuromuscular disorder that is caused by lack of α2 chain of laminin protein (LAMA2). Infants with LAMA2-CMD have poor muscle tone (hypotonia) and muscle weakness starting at birth. It affects approximately 1 in 200,000 newborns, and due to its rarity nature, there are many aspects of the disease pathophysiology that are not fully understood yet.
Using mouse model and patient-derived cells, we are trying to understand why different tissues are affected differently in this condition. Furthermore, we are developing genome editing strategies to correct the underlying mutation and upregulate a “sister” gene called LAMA1.
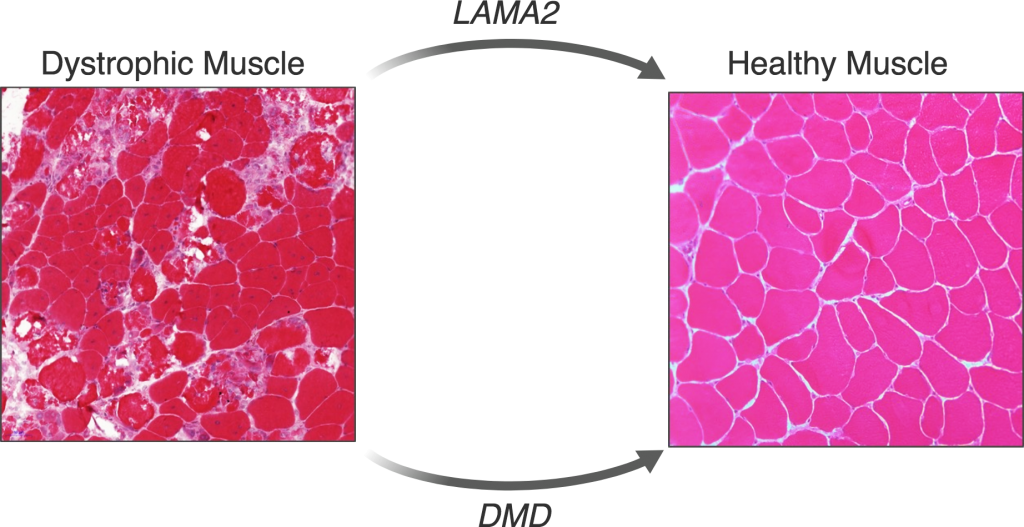
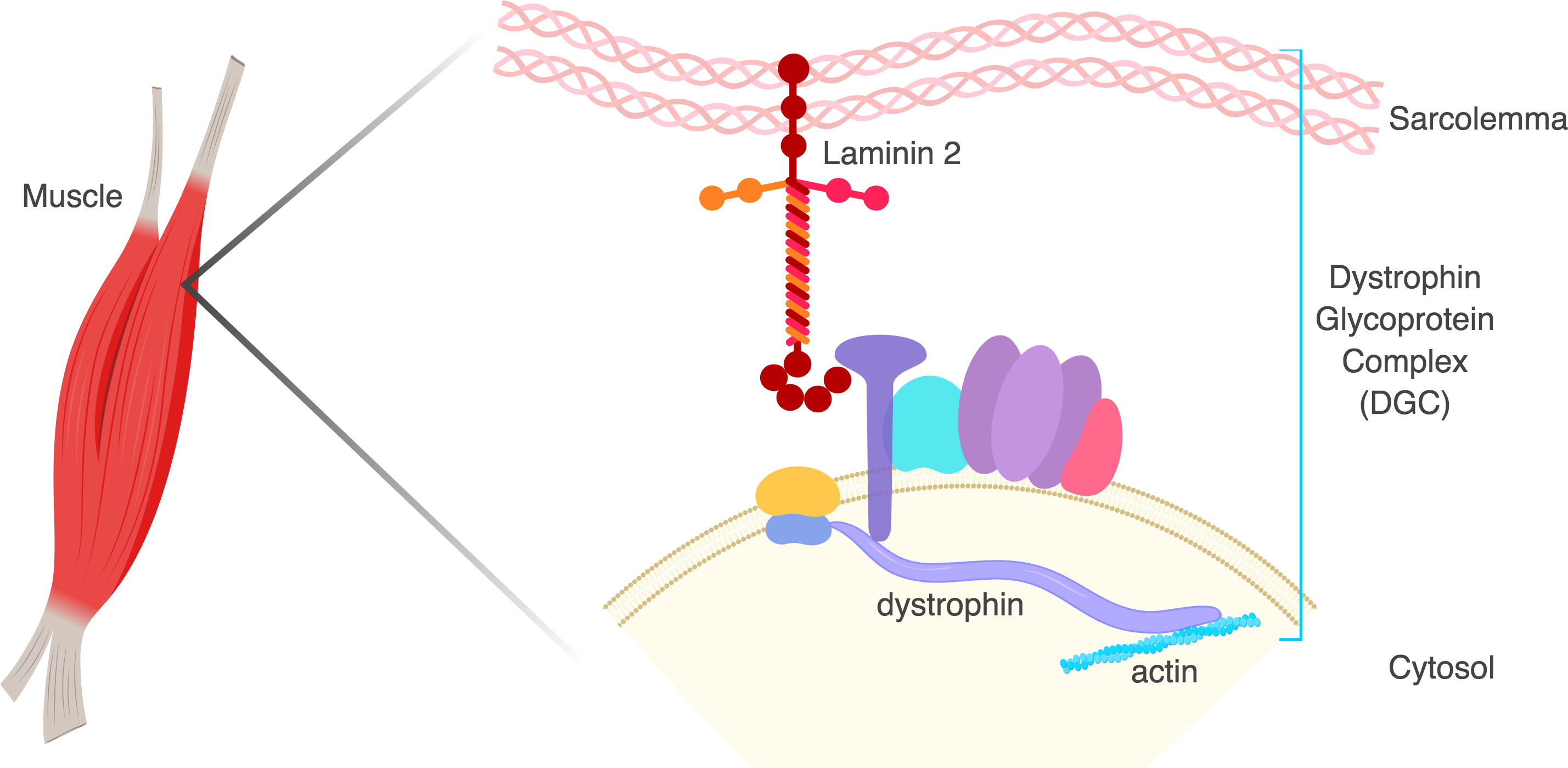
Duchenne Muscular Dystrophy (DMD)
DMD is an X-linked recessive neuromuscular disorder that affects 1 in 5,000 boys. This disease is caused by a lack of expression of the dystrophin protein, which is integral to the dystrophin glycoprotein complex (DGC) in skeletal muscle. Dystrophin creates a buffering system during cycles of muscle contraction and relaxation. Therefore the loss of dystrophin leads to progressive muscle wasting, loss of ambulation, and cardiorespiratory complications that lead to the limited life span of these DMD patients.
Our goal is to restore either truncated or full-length dystrophin using gene editing strategies to ameliorate DMD disease phenotype.
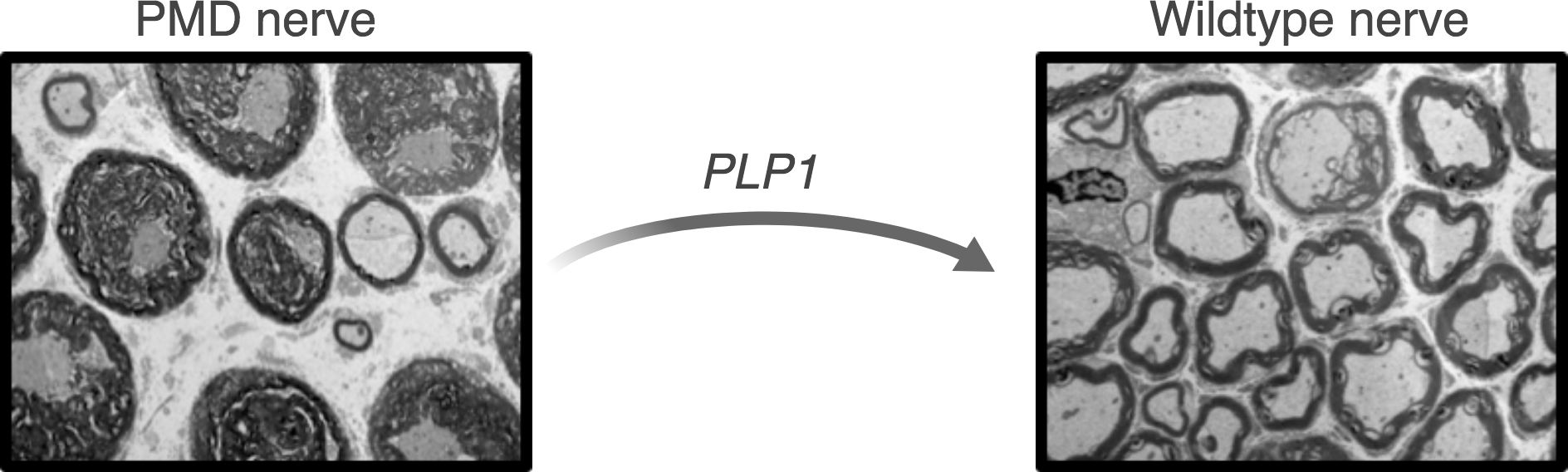
Pelizaeus Merzbacher Disease (PMD)
Pelizaeus Merzbacher Disease (PMD) is a rare X-linked pediatric leukodystrophy, that affects approximately 1 in 100,000 boys. The disease is associated with severe motor and cognitive impairment and limited life expectancy. PMD is caused by mutations in the PLP1 gene which encodes for proteolipid protein 1, one of the main components of myelin. In healthy individuals, myelin forms an insulating layer around the nerve fibers to allow fast and efficient signal conduction in the nervous system. Mutations in the PLP1 gene lead to myelination abnormalities, ultimately resulting in the onset of PMD symptoms.
This project aims to implement genome editing approaches to correct mutations causing PMD and ameliorate disease manifestation in PMD mouse models. Our objectives are to better understand the disease pathophysiology and to ultimately provide new treatment strategies for PMD patients.
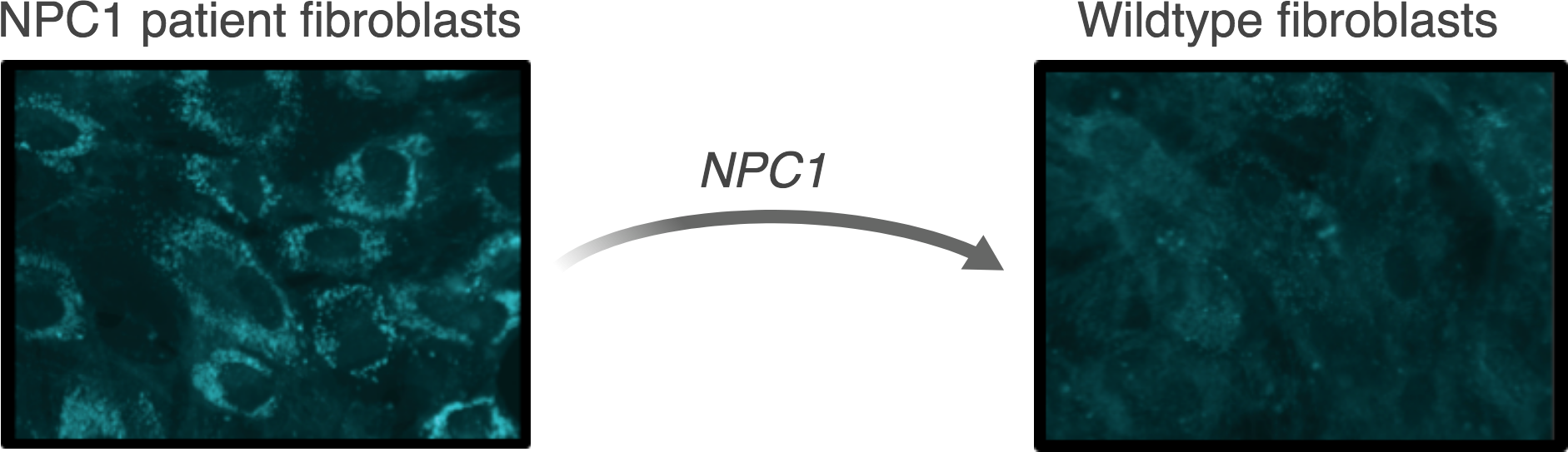
Niemann-Pick disease type C (NPC)
Niemann-Pick disease type C (NPC) is a lysosomal storage disorder that affects approximately 1 in 120,000 live births. The hallmark symptom of NPC is progressive and fatal neurodegeneration. In most cases, the disease is caused by mutations in the NPC1 gene encoding lipid transporter NPC1. These mutations cause a pathological accumulation of cholesterol and other lipids within lysosomes, which eventually lead to the fatal neurodegeneration. Presently, there is no cure for NPC, and treatment options are limited to symptom management.
We are investigating genome engineering and genome editing strategies to treat Niemann-Pick disease type C.
Joubert-like syndrome
Joubert-like syndrome is predominantly an autosomal recessive disorder, with a prevalence of approximately 1 in 100,000. It can be caused by mutations in several genes, and is part of a larger group of disorders called ‘ciliopathies’. Ciliopathies affect cellular structures called cilia, which are microtubule-based, cytoplasmic extensions which are critical for developmental and physiological functions. Joubert syndrome affects the brain and is characterized by the appearance of a typical ‘molar tooth sign’ on MRI. Therefore, affected individuals often display intellectual disability. Many other parts of the body can be affected as well, and symptoms can include weak muscle tone, abnormal breathing patterns, and abnormal eye movements.
Our goal is to investigate the significance of clinical variants in new genes and develop better therapeutic strategies for affected individuals.
STAT1 Gain-of-Function
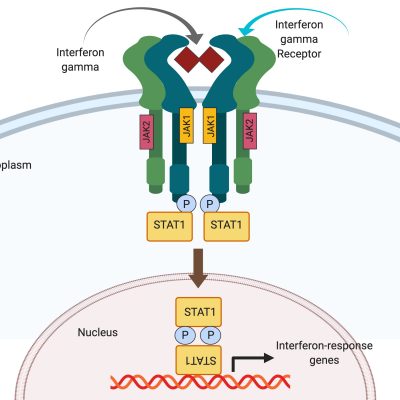
STAT1 Gain-of-Function (GOF) is a rare immunodeficiency disorder. Patients typically present as infants with recurrent fungal infections. They later develop other severe infections, as well as autoimmunity, blood vessel inflammation (vasculitis), and sometimes malignancy. Quality of life of individuals with STAT1 GOF is often poor, as they depend on life-long medications for symptoms control, spend long periods of time in the hospital, and often die in adolescence or early adulthood. Currently, treatment options for STAT1 GOF are limited to infection control and immunosuppression. Unfortunately, there is no cure for STAT1 GOF. Bone marrow transplant, which is used to treat other immunodeficiencies, is often lethal in these patients. Furthermore, the exact mechanism whereby STAT1 GOF causes disease is not entirely understood, and at present there are no adequate cell or animal models in which the disease can be studied.
The aim of our work is to create both cell and mouse models of STAT1 GOF, comparing common and severe human mutations. Such models would allow us to explore the changes that STAT1 GOF causes in the cell, and would also enable other scientists around the world to study the same disease in a standardized manner. Finally, these models have the potential to identify targets for intervention, which may be offered as therapeutic options to patients in the future.
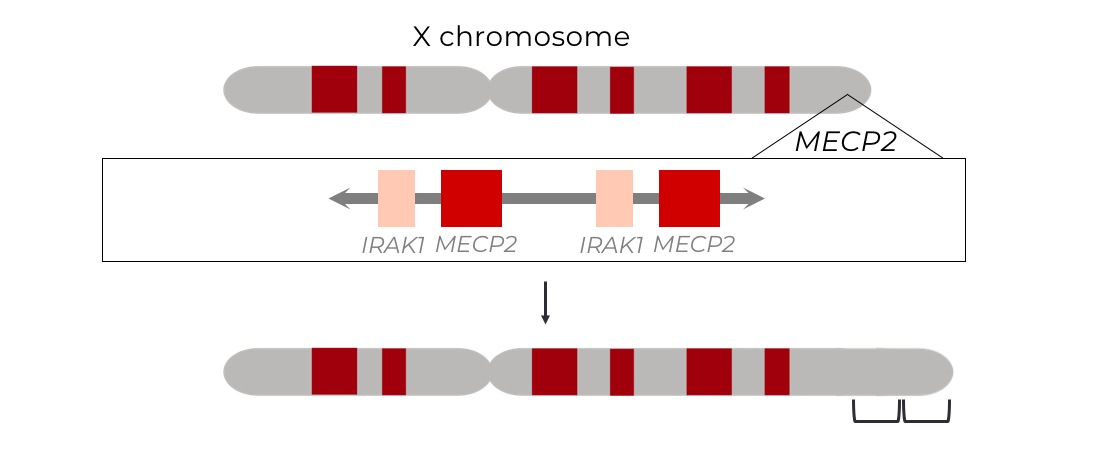
Methyl CpG Binding Protein 2 (MECP2) Duplication Syndrome
Methyl CpG binding protein 2 (MECP2) duplication syndrome (M2DS) is a rare, X-linked recessive, neurodevelopmental disorder primarily affecting young boys, and suspected to underly 1-2% of X-linked intellectual disabilities. The disease is caused by a duplication of the global transcriptional regulator, MECP2, and it’s neighboring genes, which span anywhere from 100 kb to up to 15 Mb. These duplications result in a pathological disruption of neuronal homeostasis by altering regulation of neuronal genes. This ultimately manifests in developmental delay, intellectual disability, autism spectrum disorder, and early death. Presently, there is no cure for the disease and treatment options are severely limited to symptom management.
The project aims to generate, characterize and correct patient-relevant M2DS mutations in human neuronal cells to reverse the devastating neuronal phenotypes conferred by the disease. Our objectives are to provide the crucial pre-clinical data to translate these strategies into curative clinical therapies for M2DS patients, and extend it to patients suffering from duplication disorders at large.