Research
A deeper look into our methodologies and techniques
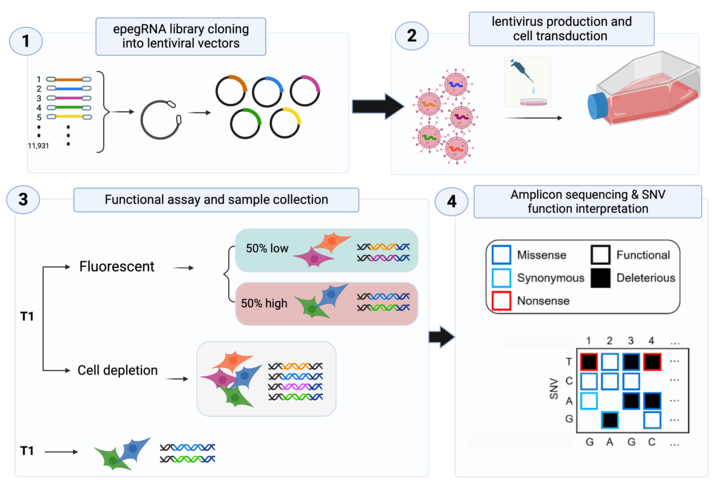
The overarching focus of the Ivakine Lab can be boiled down to two main ideas: 1) understanding the functionality of genetic variants, at scale, through saturation genome editing; and 2) developing novel CRISPR-based and small molecule-based therapeutics to generate precision therapies for rare genetic disorders — with a focus on neurodegenerative and neurodevelopmental conditions. We are using cutting edge genome editing tools and delivery vehicles to generate and correct disease-causing mutations in cell and animal models.
Development of novel therapeutic approaches for the
next generation of precision medicines
Genetic diseases manifest in various ways, some affecting the brain, muscles and several other organs. Disease-causing mutations function similarly. Our lab focuses on a variety of these mutations, including:
- simple single nucleotide changes, such as Joubert-like syndrome mutation
- small deletions and insertions, such as Tay-Sachs Disease mutation in HEXA
- duplications of large DNA fragments, such as MECP2 duplication syndrome
- deletions of large genetic regions, such as the exon 52 deletion in the gene that causes Duchenne Muscular Dystrophy
A plethora of disease causing mutations, coupled with different organs’ involvement, makes it challenging to develop therapies. We are using a whole arsenal of recently developed genome engineering tools (base editing, prime editing) to first model a mutation in a relevant cell line, analyze it for functional consequences and interrogate various correction strategies. We then develop a relevant model of a disease, and after thorough characterization, we apply our most promising strategies to elicit a therapeutic effect in vivo. We are closely working with regulatory agencies to bring these novel therapies into the clinic.
CRISPR and drug-based screens to rescue disease pathogenesis
Image credit: HHMI BioInteractive
We have recently initiated CRISPR-KO and CRISPRa screens to not only gain a deeper insight into disease pathogenesis but to also identify molecular pathways in which modulation may influence disease progression and severity. Complementary, high throughput functional assays are also performed to identify novel therapeutic candidates.